Epigenetics
Following the completion of the Human Genome Project, much of Biology’s focus has shifted from the DNA sequence of genes to their regulation in response to environment, in which epigenetics represents a core mechanism. The term Epigenetics was first defined in early 20th century by the British biologist Conrad H. Waddington as a layer of mechanisms that resides above the level of the genes, which control their output in order to specify cell fate determination during organismal development that from the very same genetic information gives rise to functionally different cell types e.g. a neuron versus a hepatocyte. However, it is now clear that these mechanisms operate beyond organismal development and that experience, be it environmental stimuli, maternal behavior, psychological or physical stress, learning or exposure to drugs of abuse, leads to active regulation of the chemical and three-dimensional structure of DNA in the nervous system, i.e. that experience regulates epigenetic mechanisms in the CNS. In this postgenomic era, there is no longer a mechanistic dichotomy between nature and nurture, but rather a dynamic interplay between genes and experience through a biochemically driven mechanistic interface - That is epigenetics.
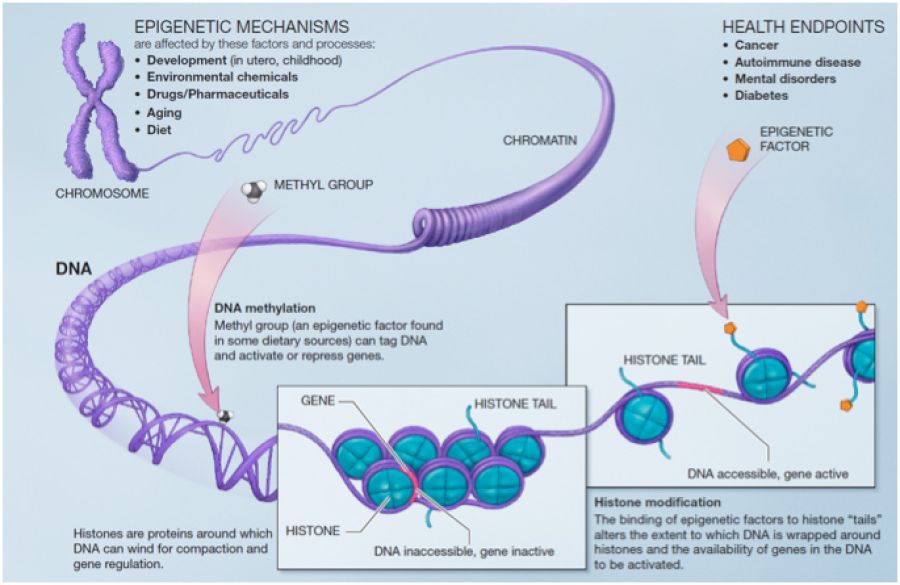
Epigenetic mechanisms
The major epigenetic mechanisms include DNA methylation, histone modifications and nucleosome and higher order chromatin remodeling, ncRNAs (non-coding RNAs) and RNA editing. DNA methylation refers to covalent modification of cytosine residues to form 5-methylcytosine (5-mC). The presence of 5-mC at specific gene loci was previously thought to promote transcriptional repression and long-term gene silencing. However, based on recent discoveries it is now clear that cytosine methylation can also be associated with transcriptional activation. Besides DNA cytosine methylation, other chemical modifications of cytosine in DNA have recently been documented to exist, including 5-hydroxymethylcytosine (5-hmC) formation and methylcytosine oxidation to generate 5-formylcytosine and 5-carboxylcytosine. The functional roles of these novel modifications are not fully established, yet they appear to be particularly prominent in the nervous system and in the zygote; two highly plastic tissues in the body. This is a hot area of investigation in the field at present time. Histone modifications are the second major category of epigenetic biochemical mechanisms in cells. DNA is wrapped around a histone protein octamer, forming a fundamental chromatin structure, the nucleosome. Each nucleosome contains 2 of each core histone protein (H2A, H2B, H3, and H4). Histone posttranslational modifications that have functional consequences on gene readout are multitudinous, including lysine acetylation, lysine mono/di/tri-methylation, arginine mono/di-methylation, serine/threonine phosphorylation, histone monoubiquitination and histone poly ADP-ribosylation, and they regulate the nucleosome structure in order to modulate transcriptional readout of the associated gene. Individual isoforms of histone monomers can also be swapped in and out of the octamer, a regulatory mechanism referred to as histone subunit exchange. Collectively, these mechanisms have given rise to the concept of a histone code, wherein histone modifications are interpreted as a combinatorial code regulating gene transcription rates at specific loci across the genome. Nucleosome and higher-order chromatin remodeling is a third layer of epigenetic mechanisms that are driven by macromolecular complexes containing various proteins such as SWI/SNF, Polycomb and Trithorax groups with the ability to simultaneously read, erase, and write epigenetic marks to promote remodeling. Newly recognized classes of ncRNAs include microRNAs (miRNAs), endogenous short-interfering RNAs, PIWI-interacting RNAs, small nucleolar RNAs and long ncRNAs. MicroRNAs, which engage in posttranscriptional gene regulation, are the best-characterized class of ncRNAs. Long ncRNAs (lncRNAs) on the other hand are the most abundant but least well-characterized class of ncRNAs and are believed to have novel activities such as recruiting transcriptional and epigenetic regulators to specific genomic loci and local protein synthesis at synapses. Last but not least, RNA editing is a mechanism for modifying RNA molecules. For instance, RNA editing can change codons in mRNAs, leading to expression of protein molecules different from those encoded in the genome.
Neuroepigenetics: State-of-the-art and our research program
Over the last decade, there has been a great expansion of the number of research articles published concerning epigenetic mechanisms in the nervous system. While much of the intricacy surrounding epigenetic mechanisms is yet to be defined fully, epigenetics has already revolutionized our understanding of how the human brain evolved and how neural cellular diversity, connectivity, plasticity and intellectual abilities emerge. Not surprisingly, it is also becoming apparent that epigenetics is intimately involved in brain disease pathogenesis. The contributions of epigenetic abnormalities to brain disease pathophysiology range from being primary pathogenic mechanisms, e.g. MECP2 gene mutations being the primary causes of Rett syndrome, to differential epigenetic profiles linked to aberrant gene expression programs, e.g. in Huntington’s disease, promoting the development of paradigm-shifting clinical applications. Our previous work has shown altered histone acetylation patterns in brain after stroke and identified certain members of histone acetylation machinery as part of endogenous protection programs in the brain. Targeting histone acetylation by histone deacetylase inhibitors, via enhanced expression of neuroprotective effector genes, led to potent therapeutic effects in stroke models. In Huntington’s disease (HD), we measured epigenetic changes such as histone acetylation and methylation, and DNA methylation in cell and mouse models at early and late disease stages and identified specific transcription factors that were linked to aberrant gene expression in HD. Chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq) for histone modifications has led to a striking discovery of a distinct chromatin signature which identifies the promoters of neuronal genes that are down-regulated in HD brain, suggesting a specific regulatory mechanism for the genes that are targeted by mutant Huntingtin gene in mouse as well as human neurons.
The focus of our group is to understand the epigenetic, transcriptional and posttranscriptional regulatory mechanisms of neuronal gene expression in brain disease. By applying next generation DNA sequencing approaches to relevant mouse models and patient cells combined with neuropathological, mouse brain imaging and behavioral analyses, our major goal is to characterize gene expression and epigenetic changes throughout the genome in vulnerable neurons and establish links to neuronal dysfunction and manifestation of disease phenotypes. Our current research efforts are organized into three main programs.
I. Profiling epigenetic and transcriptional mechanisms of neuronal vulnerability in disease
Many neurological and psychiatric diseases are characterized by the early dysfunction of selective populations of neurons in the brain, followed only later by more widespread dysfunction and degeneration. Understanding the cause of the enhanced vulnerability in any given disease could be a key to unravel the true cause of pathogenic demise and lead to novel therapeutic targets. We and others have shown that large number of genes are changed in their expression and epigenetic regulation in Huntington’s disease (HD) brain in experimental models as well as patient postmortem brains and induced pluripotent stem cells (IPSCs) from patients. However, due to the use of bulk tissues in previous studies, it is unknown whether the key changes in the vulnerable neurons were truly represented in these results or obscured by mixture of the different cell types of brain. By utilizing current cell-type specific methodologies, e.g. reporter mouse lines, our goal is to profile those neurons that show early and selective dysfunction in HD. We use next generation sequencing techniques to measure genome-wide changes in gene expression and epigenetic marks. For assessing the neuropathological and behavioral correlates of the measured molecular changes, we utilize immunohistochemistry and behavioral tests in mouse models. These studies aim at identifying differential epigenetic landscapes, associated gene expression changes and transcriptional/epigenetic regulators that are specific to vulnerable neuronal populations, whose altered activities may underlie the behavioral deficits in the HD mouse. In close collaboration with Josef Priller (Charité Berlin, Department of Neuropsychiatry), we use mortem human brain tissues from HD patients and patient IPSCs for evaluating the concordance of pathogenic events in mice and human.
II. Pre-clinical testing of new therapeutic targets
Our goal is to test the therapeutic potentials of the key transcriptional/epigenetic targets that are identified by the afore-mentioned profiling studies to develop effective treatments for brain disease. This could be achieved by targeting the altered epigenetic/transcriptional regulators as well as interactions with partnering co-regulators and/or upstream signaling pathways. We work towards this goal by using two main strategies: i) Targeting of the epigenome by specific agents. We and others across the globe have previously reported potent neuroprotective effects of various histone deacetylase (HDAC) inhibitors and DNA methylation targeting agents against an array of brain diseases, including stroke and Huntington’s disease. As a result of rigorous efforts in recent years, there are now more specific HDAC inhibitors and other chromatin targeting agents available. We are currently testing some of these recently-developed, specific epigenome targeting agents in HD and other neuropsychiatric disease models. ii) Gene therapy. For specific targeting of genes, we use viral vectors, delivered by stereotaxic injections into mouse brain, to enhance or to reduce the expression of transcriptional/epigenetic regulators in defined brain areas. For outcome measurement, in both the pharmacological and gene therapy approaches, we utilize immunohistochemistry, small animal MRI and a battery of behavioral paradigms for emotional, cognitive and motor disease phenotypes. Findings from these studies will potentially promote the development of new therapeutic interventions in neuropsychiatric disease.
III. Linking gene expression programs to neuronal function and morphology
We have established an in vitro model of HD that includes the N-terminus of the mutated Huntingtin (mut HTT) gene and is based on lentivirus – vector mediated transduction of primary mouse neurons. The model mimics several key features observed in HD mouse models and patients like prominent nuclear inclusions and dysregulation of key genes in both cortical and striatal neurons. In close collaboration with Christian Rosenmund (Charité Berlin, Department of Neurophysiology), we study the effects of Huntingtin (HTT) mutation on neuronal morphology, synapse formation and function, and gene expression at single neuron level. Electrophysiological measurements for various synapse function parameters, e.g. IPSCs, EPSCs and readily releasable vesicles, are followed by single cell RNA-Seq. For assessments of neuronal morphology, e.g. soma size, dendrite length and synapse density, immunocytochemistry for relevant proteins is utilized. Our ongoing work includes application of single cell RNA-seq to disease neuronal cultures, such as HD and Rett, following electrophysiological measurements in experiments that aim at assessing beneficial effects of therapeutic candidates.